qgisprocess package created by Dewey Dunnington is a wonderfull package that allows interacting with QGIS directly from R. The package provides very nice, but a little bit low level interface to qgis_process commandline tool. My only problem with the package is that user needs to be rather familier with QGIS tools and their arguments to be able to effectively use this package.
Looking at the qgisprocess I quickly realized that it should be possible to utilize this package to generate R code and roxygen help texts for individual QGIS tools. After finding this post, which pretty much covers how to handle this issue, I dedicate some time to putting it together. The result is qgis package. The package contains something almost 1000 functions for native QGIS provide GDAL, GRASS and SAGA algorithm provides. This version is built online using Github Actions against latest QGIS release (3.24 when I am writing this). However, the package can be build locally (as described here) and in that case it will also generate functions for providers from instaled plugins, scripts and models. The local package build takes significant amount of time - around 30 minutes for basic installation. But such installation allows user to interact with the full QGIS setup that he is normally using.
The package provides functions with templete name {algorithm-provider}_{algorithm-id} (i.e. qgis_buffer or grass7_v_buffer), so that it is obvious what is algorithm provider and its name. This allows for rather convinient code completation in RStudio.
The package can be installed directy from Github using remotes package:
# install.package("remotes")
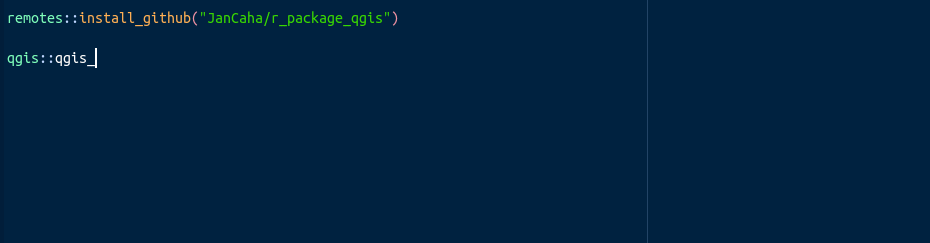
remotes::install_github("JanCaha/r_package_qgis")
Its probably not ideal to load the package with such large number of functions explicitly into R. Instead the functions should be called as:
# install.package("remotes")
qgis::qgis_buffer()
which should be much more convient for practical use.
If you are inside RStudio then this will give you parameter name suggetion and autocompletion as well as tool help that is translated into R style (which R users are familier with) from QGIS style (which looks slightly different).
Two screen captures from RStudio showing the the autocompletation and help:
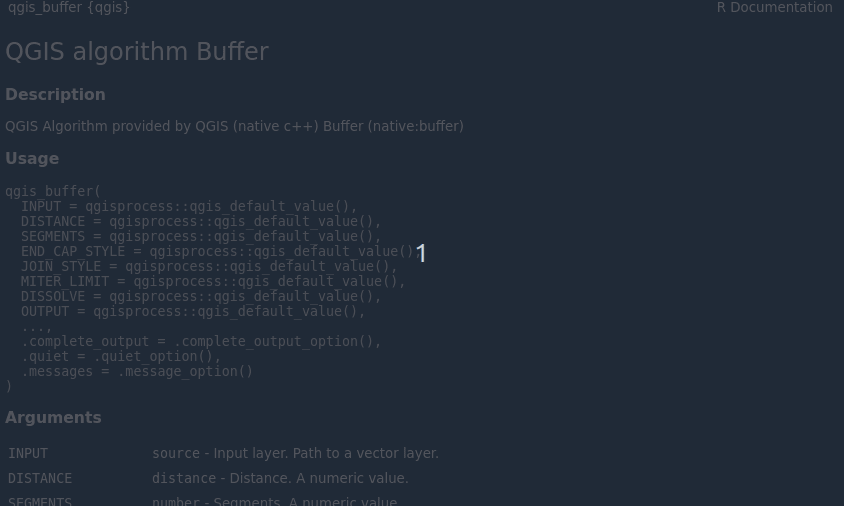
One interesting difference came up from this issue. If you need to pass flags (parameters starting with -) to GRASS algorithms, they need to be wrapped inside backtics `-m`=TRUE so that qgisprocess interprets it correctly. In qgis these parameters are renamed to start with . e.i. .m, so they need not to be wrapped in backticks. The presence of help also allows you to quickly check what values should these flags be set to.
So these two calls are equal:
dem_wetness <- qgis_run_algorithm("grass7:r.geomorphon",
elevation = dem,
`-m`="true")
dem_wetness <- qgis::grass7_r_geomorphon(elevation = dem, .m = TRUE)
The first one relyies on knowledge of algorithm provider, name and its arguments, the second can be found much more easily and provides full access to help and autocompletation.
The simple example of use can look like this:
library(sf)
fname <- system.file("shape/nc.shp", package="sf")
nc <- st_read(fname)
buffered <- qgis::qgis_buffer(INPUT = nc,
DISTANCE = 0.5,
END_CAP_STYLE = "Flat") %>%
st_as_sf()
plot(buffered)
Help for all tools and addition information is summarized in: